
DECODING SCIENCE POST by Caitlin J. Curry from The Conversation
Zambia, a country in southeast Africa, has approximately 1,200 lions, one of the largest lion populations on the continent. More than 40% of the U-shaped country is protected land, with over 120,000 square miles of national parks, sanctuaries and game management areas for lions to roam.
Zambian lions are split into two subpopulations, with one in the Greater Kafue Ecosystem in the west and the other in the Luangwa Valley Ecosystem in the east. Between these two geographically different regions lies Lusaka, Zambia’s largest city, which is surrounded by farmland.
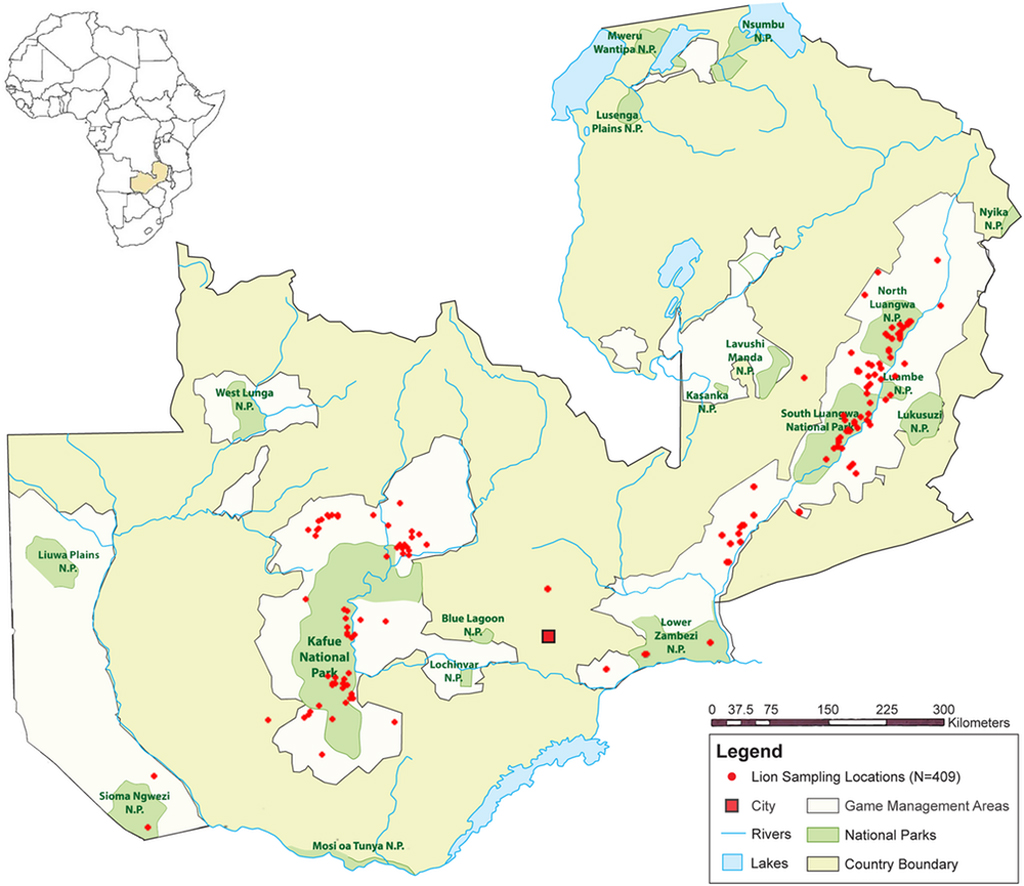
People had assumed that the two groups of lions did not – even could not – mix. After all, they’re separated by a geographical barrier: the two regions feature different habitats, with the east an offshoot of the Great Rift Valley system and the west part of the southern savannas. The lions are also separated by what’s called an anthropogenic barrier: a big city that lacks wildlife protection, making it seemingly unsuitable for lions.
So my colleagues and I were surprised when we found that a small number of lions are in fact moving across the area in between presumed to be uninhabitable by lions. These sneaky lions – and their mating habits – are causing the high levels of genetic diversity we found in the entire Zambian lion population.
Identifying which genes are where
Working with the Zambian Wildlife Authority, biologist Paula White collected hundreds of biological samples from lions across Zambia between 2004 and 2012. Eventually, a box of this hair, skin, bone and tissue, meticulously packaged and labelled with collection notes and sampling locations, arrived at my lab at Texas A&M University.

Our goal was to investigate genetic diversity and the movement of various genes across Zambia by extracting and analysing DNA from the lion samples.
From 409 lions found inside and outside of protected lands, I looked at two kinds of genes, mitochondrial and nuclear. You inherit mitochondrial DNA only from your mom, while you inherit nuclear DNA from both of your parents. Because of these differences, mitochondrial and nuclear genes can tell different genetic stories that, when combined, paint a more complete picture of how a population behaves.

My mitochondrial analysis verified that, genetically, there are two isolated subpopulations of lions in Zambia, one in the east and one in the west. However, by also looking at the nuclear genes, we found evidence that small numbers of lions are moving across the “unsuitable” habitat. Including nuclear genes provided a more complex picture that tells us not only which lions were moving but also where.

Genes on the move as lions roam
The amount of variation from alternate forms of genes found within a population is known as genetic diversity. Genetic diversity is important for a wildlife population because more genetic options give animals a greater chance for adaptation in a changing environment. Genetic diversity can also tell biologists about ways a population can fluctuate.
To a geneticist, migration, also referred to as gene flow, is the movement of genes from one geographical place to another. Mitochondrial DNA, inherited from the mother, can only tell researchers where genes from mom have been.
In the lion mating system, males travel long distances to find new prides, while females remain in or close to the pride they were born in. So, for the lion, it’s primarily males that are responsible for the movement of genes between prides. This male-mediated gene flow explains the lack of gene flow seen in mitochondrial genes compared to that of nuclear genes – female lions aren’t making the journey, but they do mate with new males who come from far away.
Male-mediated gene flow has helped keep the lions of Zambia genetically healthy, increasing genetic diversity by introducing new genes to new areas as male lions move between subpopulations. The eastern and western subpopulations each have high levels of genetic diversity; since only a few lions move between the groups each generation, the subpopulations stay genetically distinct.
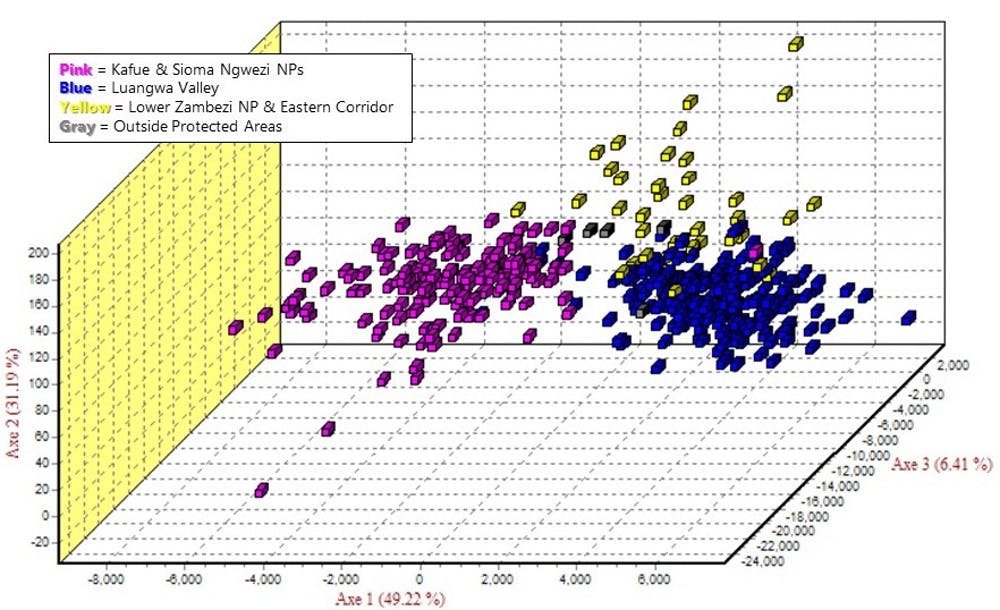
My colleagues and I were also able to determine where the lions are moving based on which individuals are more genetically similar to each other. Lions in the North and South Luangwa National Parks, part of the eastern subpopulation, appear entirely separated from the western subpopulation. Gene flow is occurring through the southern regions of the east subpopulation.
Lions are most likely travelling a route between the Lower Zambezi National Park and eastern corridor to the Kafue National Park in the west, possibly along the Kafue River. We can’t tell which way they’re moving, but by looking at where lions are more closely related, we can see where genes are being moved.
Lion data can help manage wildlife overall
Human-lion conflict is a big issue in Zambia, particularly outside of protected land. If lions were moving across human-dominated areas, you’d think they’d be seen and reported. But these lions are sneaking through virtually undetected – until we look at their genes.
As a large, charismatic carnivore, lion research and conservation influences many other species that share their habitat.
Wildlife managers can use these findings to help with lion conservation and other wildlife management in and around Zambia. Now that we generally know where lions are moving, managers can focus on these areas to find the actual route the big cats are taking and work to maintain or even increase how many lions can move across these areas. One of the ways of doing this is by creating more protected land, like corridors, to better connect suitable habitat.
Full report: Caitlin J. Curry, Paula A. White, James N. Derr (2019). Genetic analysis of African lions (Panthera leo) in Zambia support movement across anthropogenic and geographical barriers. PLOS ONE. https://doi.org/10.1371/journal.pone.0217179
To comment on this story: Login (or sign up) to our app here - it's a troll-free safe place 🙂.![]()






